5D Navigator:
Import Confocal/Lightsheet Images
BIOQUANT opens image data from confocal or light sheet microscopes into the 5D Image Navigator.
The 5D Image Navigator supports the OME-TIF (.OME.TIF) image format, for single images or sequence of image up to 4 GB. If your confocal or light sheet microscope does not save directly to OME-TIF use the free BIO-FORMATS plug-in to ImageJ to convert.
Like the Image Navigator, the 5D Image Navigator acts as a virtual microscope. Zoom out to see the entire image, or in to a high resolution field.
In addition, the 5D Image Navigator provides fluorescence channel mixing and depth of field adjustment, as well as Z plane navigation.
An Introduction to 5D Imaging
5D imaging is a strategy for organizing images of fluorescent samples. The 5 dimensions we’re talking about are x width, y height, c fluorescent channel, z focal plane, and t time point.
The simplest images is an area of a sample with a single fluorescent label at a single focal plane at a particular time. As you add more fluorescent channels, add more Z planes, and add more time points, the number of dimensions expands from 2 to 5. The following series of diagrams helps to explain.
DETAILS OF 5D IMAGING: A 2D IMAGE
A single fluorescent label at a single focal point at a particular time creates a 2D image.
DETAILS OF 5D IMAGING: A 3D IMAGE
Three fluorescent labels at a single focal point at a particular time create a 3D image.
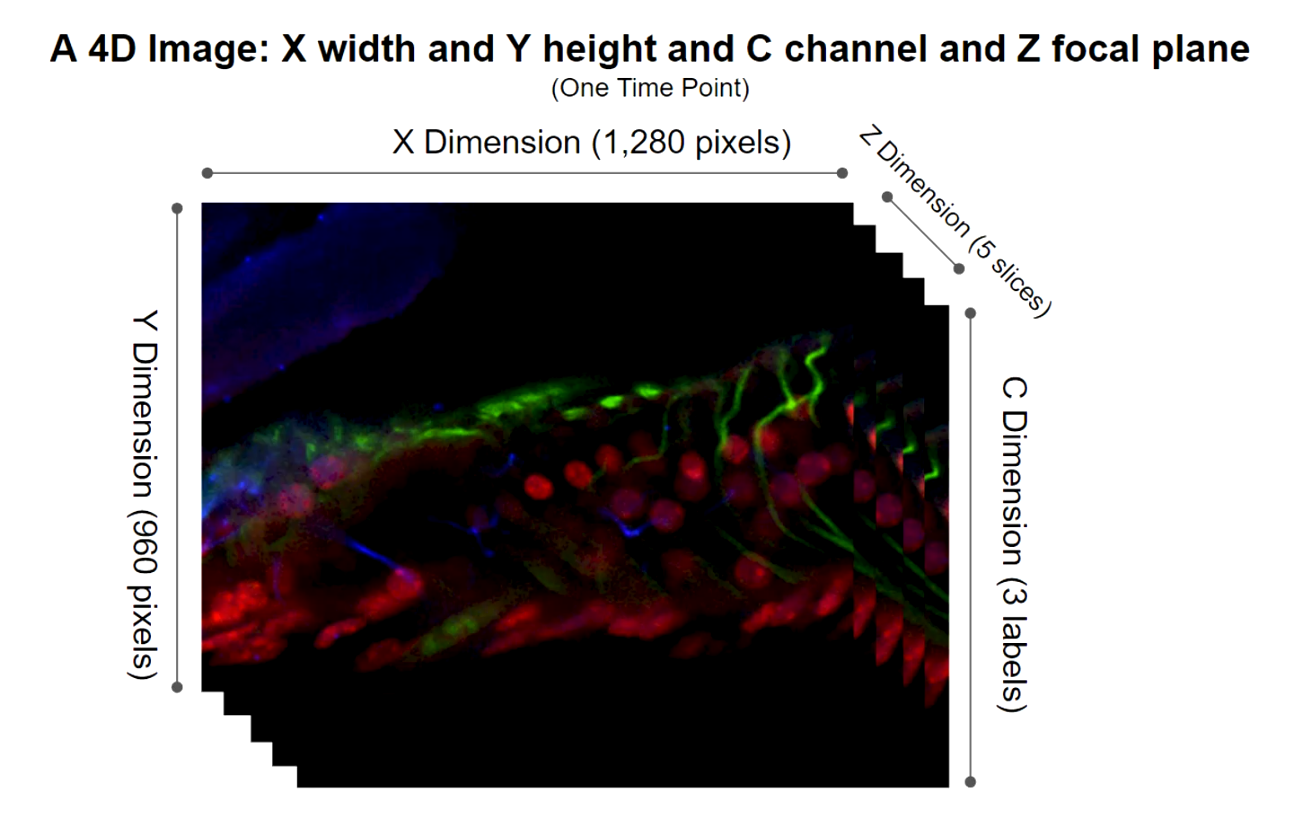
DETAILS OF 5D IMAGING: A 4D IMAGE
Three fluorescent label at a five focal points at a particular time creates a 4D image.
DETAILS OF 5D IMAGING: A 5D IMAGE
Three fluorescent label at a five focal points at 3 distinct time points creates a 5D image.
Confocal and lightsheet microscopes can collect many images of a sample each representing different combinations of XYCZ, and T. These images can easily reach multiple gigabytes in size. To simplify moving all that data around, a few different solutions have emerged.
First, confocal and lightsheet companies made their own proprietary file formats.
Then, ImageJ created Hyperstacks using the TIF format. Hyperstacks are a way to store 5D data in TIF files. TIF was an easy choice because the TIF file standard actually supports storing many different images in a single file.
What was missing from ImageJ’s approach was a more complete way to store the necessary information describing all the various images in the TIF file. So the folks at the Open Microscopy Environment project created OME-TIFF.
OME-TIFF uses the basic idea of storing multiple images in a single TIF file, but adds a formal framework for storing all the necessary information about those images in the same TIF file. This information is called metadata.
The BIOQUANT 5D Image Navigator supports the OME-TIFF format.
We chose this format because the Open Microscopy Environment project also made a free ImageJ plugin called BIO-FORMATS which can convert just about every other 5D format into OME-TIFF. That allows BIOQUANT to support the broadest array of 5D data.
The OME-TIFF Format
Multi-dimensional image data in the OME-TIFF format is loaded into the 5D Navigator using with the new “Open OME TIF File to 5D Image Navigator” menu item on the Sequence button in the Image region.
If your confocal or lightsheet software cannot save to OME-TIFF, the BIO-FORMATS plug-in to ImageJ can be used to convert it to the OME-TIFF format. We have a procedure that walk you through using ImageJ to covert to OME-TIFF, contact us!
The BIOQUANT 5D Image Navigator supports both single file and multiple file OME-TIFF files, up to 4 GB in total size.
When loading 5D data into BIOQUANT, you’ll see the OME-TIFF metadata displayed on a configuration box. This gives you chance to double check that the data are accurate and add any missing data.
5D IMAGE PARAMETERS
Displays OME-TIF metadata.
Fluorescence Channel Mixing and Depth of Field Adjustment
Assignments can be made for different fluorescent labels (channels) to different colors.
By default, the first channel is set to red, the second to green, and the third to blue. Channels can be assigned to the preferred color. If there are more than 3 channels in the image data, only three can be visualized at one time.
Adjustment of the depth of field in the Image Window.
By default, the Image Window displays only the current Z position on the 5D Image Navigator Z position slider. But the depth of field can be expanded using a maximum intensity algorithm. Increase the Expand Depth of Field value to indicate how many additional Z planes above and below the current position should be merged into the image.
5D IMAGE NAVIGATOR - CORTICAL BONE INNERVATION.OME.TIF*
In this example, channel 2, which labels one kind of nerve, and is mapped to Red.
Z Plane Navigatrion
5D IMAGE NAVIGATOR - CORTICAL BONE INNERVATION.OME.TIF*
In this example, there are 121 Z Planes.
The OME-TIFF images often contain multiple Z Planes. The Z Plane slider on the 5D Image Navigator is used to sequence through the Z planes. The Go To box is used to go to a specific Z plane.
The Combine button is used to combine all Z planes or a subset of Z planes into a single image using a maximum intensity projection algorithm.
Once the final image is in the achieved, you can measure data as usual in BIOQUANT. Or you can save the image to share with others or measure on a BIOQUANT Satellite System.
* CORTICAL BONE INNERVATION.OME.TIF
Multi-channel Confocal Stack of Cortical Bone Innervation by Erica Scheller and Washington University in St. Louis (https://sites.wustl.edu/schellerlab/) is licensed under CC BY 4.0. To view a copy of this license, visit https://creativecommons.org/licenses/by/4.0.